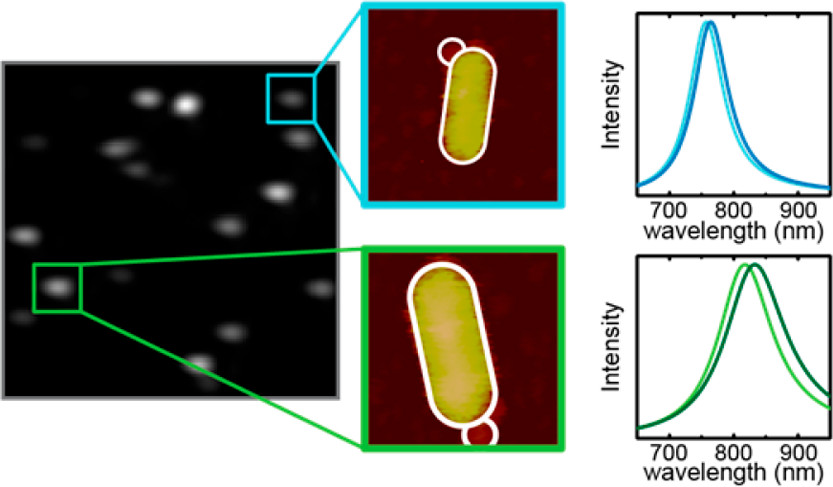
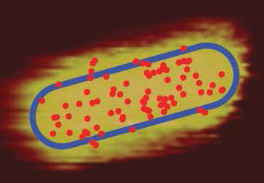
The high sensitivity of localized surface plasmon resonance sensors to the local refractive index allows for the detection of single-molecule binding events. Though binding events of single objects can be detected by their induced plasmon shift, the broad distribution of observed shifts remains poorly understood. Here, we perform a single-particle study wherein single nanospheres bind to a gold nanorod, and relate the observed plasmon shift to the binding location using correlative microscopy. To achieve this we combine atomic force microscopy to determine the binding location, and single-particle spectroscopy to determine the corresponding plasmon shift. As expected, we find a larger plasmon shift for nanospheres binding at the tip of a rod compared to its sides, in good agreement with numerical calculations. However, we also find a broad distribution of shifts even for spheres that were bound at a similar location to the nanorod. Our correlative approach allows us to disentangle effects of nanoparticle dimensions and binding location, and by comparison to numerical calculations we find that the biggest contributor to this observed spread is the dispersion in nanosphere diameter. These experiments provide insight into the spatial sensitivity and signal-heterogeneity of single-particle plasmon sensors and provides a framework for signal interpretation in sensing applications.
- March 12, 2018
- Uncategorized
- Peter Zijlstra
Related Articles
Research paper published in JPC Letters
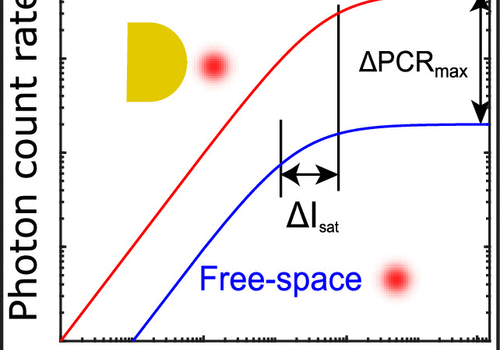
DOI: doi.org/10.1021/acs.jpclett.0c00155 Yuyang Wang, Matěj Horáček, and Peter Zijlstra Plasmon resonances have appeared as a promising method to boost the fluorescence intensity of single emitters. However, because research has focused...
Research paper published in JPCC
All-Optical Imaging of Gold Nanoparticle Geometry Using Super-Resolution Microscopy Adam Taylor, Rene Verhoef, Michael Beuwer, Yuyang Wang, and Peter Zijlstra DOI: 10.1021/acs.jpcc.7b12473 We demonstrate the all-optical reconstruction of gold nanoparticle...
Rachel wins The Best Paper Award at Europtrode 2018
The Europtrode XIV in 2018 was held on 25-28th March in Naples, Italy. Rachel's talk on plasmonic biosensors won The Paper Award. Congratulations to Rachel! The Best Paper Award is...