Plasmon-Enhanced Single-Molecule Enzymology
Yuyang Wang and Peter Zijlstra
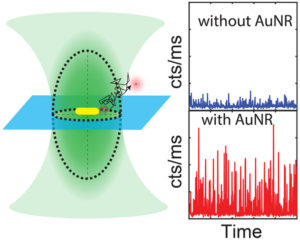
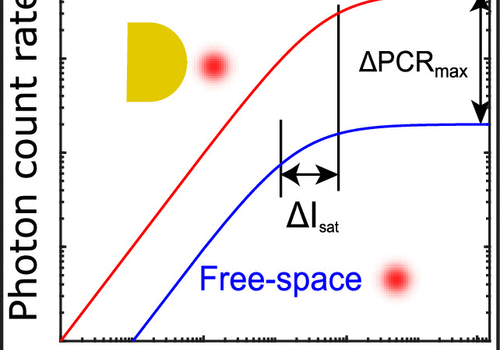
We present a numerical study on plasmon-enhanced single-molecule enzymology. We combine Brownian dynamics and electromagnetic simulations to calculate the enhancement of fluorescence signals of fluorogenic substrate converted by an enzyme conjugated to a plasmonic particle. We simulate the Brownian motion of a fluorescent product away from the active site of the enzyme, and calculate the photon detection rate taking into account modifications of the excitation and emission processes by coupling to the plasmon. We show that plasmon enhancement can boost the signal-to-noise ratio (SNR) of single turnovers by up to 100 fold compared to confocal microscopy. This enhancement factor is a trade-off between the reduced residence time in the near-field of the particle, and the enhanced emission intensity due to coupling to the plasmon. The enhancement depends on the size, shape and material of the particle and the photophysical properties of the fluorescent product. Our study provides guidelines on how to enhance the SNR of single-molecule enzyme studies and may aid in further understanding and quantifying static and dynamic heterogeneity.
Related Articles
Frank Bloksma, Peter Zijlstra* DOI link: https://pubs.acs.org/doi/abs/10.1021/acs.jpcc.1c06665 Colloidal plasmonic materials are increasingly used in biosensing and catalysis, which has sparked the use of super-resolution localization microscopy to visualize processes at...
Original post can be found on the TU/e website. photo Vincent van den Hoogen Peter Zijlstra recently received a Consolidator Grant of € 2 million from the European Research Council...
DOI: doi.org/10.1021/acs.jpclett.0c00155 Yuyang Wang, Matěj Horáček, and Peter Zijlstra Plasmon resonances have appeared as a promising method to boost the fluorescence intensity of single emitters. However, because research has focused...